Download DiMmer here.
In order to run DiMmer, your computer must be able to run Java 8 applications. You can download Java here.
In order to run DiMmer, use the console of your operating system (Linux/Mac: bash or similar, Windows: cmd) and navigate to the selected download folder and type
java -Xmx4096M -jar dimmer.jar
This is necessary as the Methylation dataset exceed the standard values for the Java virtual machine. The parameter Xmx is used for increasing the allowed memory usage of DiMmer. In case DiMmer still aborts with a out-of-memory message, try increasing the value accordingly.
Jllumina
Download the Jlummina java library here. Jlummina is the core library for processing Illumina methylation reads and is the core of DiMmer. This library supports low level manipulation of probes from the Infinium HumanMethylation450 BeadChip Kit and the Infinium MethylationEPIC BeadChip . Besides the capability of parsing Illumina IDAT files, the library also provides methods for quality control, normalization, cell composition estimation, and statistical analysis (t-test, regression, permutation test, correction for multiple testing).
// Reading a single idat file
ReadIDAT gIdat = new ReadIDAT();
gIdat.readNonEncryptedIDAT("9969489068_R01C01_Grn.idat");
For a full code example consider this file. For access the API reference click here
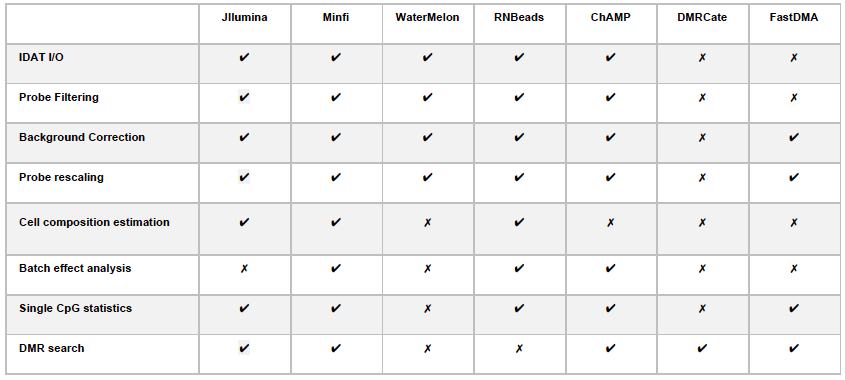
The capabilities of Jllumina compared to the other R libraries can be visualized bellow:
Resources
A case-control test data set can be downloaded here. This data set can be used for easily testing DiMmer and its capabilities, like the identification of differentially methyalted CpGs and the discovery of differentially methylated regions. The zip file contains the IDAT files of a Infinium HumanMethylation450 BeadChip Kit.
The IDAT files is only the first part of DiMmer's input. Additionally, you have to provide a sample annotation file which provides meta information of your experiment and links the different probes to the corresponding IDAT file. The file format is very simple, it is just a comma-separated file with at least wo columns linking the probe to the IDAT file: Sentrix_ID and Sentrix_Position. Optionally, you can provide three more standard columns: Group_ID, Pair_ID and Gender_ID. Besides these standard columns, you can add an arbitrary amount of columns with additional columns describing your experiment, for instance age, weight, etc.
Click here to download the sample annotation file accompanying our test dataset.